what does it mean for dna to be complementary
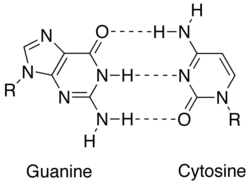
Match upwardly between two Dna bases (guanine and cytosine) showing hydrogen bonds (dashed lines) holding them together

Match up between ii Dna bases (adenine and thymine) showing hydrogen bonds (dashed lines) property them together
In molecular biology, complementarity describes a human relationship between two structures each following the lock-and-key principle. In nature complementarity is the base of operations principle of DNA replication and transcription as it is a holding shared between 2 Dna or RNA sequences, such that when they are aligned antiparallel to each other, the nucleotide bases at each position in the sequences will exist complementary, much like looking in the mirror and seeing the reverse of things. This complementary base pairing allows cells to copy information from one generation to another and fifty-fifty detect and repair impairment to the information stored in the sequences.
The degree of complementarity betwixt ii nucleic acid strands may vary, from complete complementarity (each nucleotide is across from its opposite) to no complementarity (each nucleotide is not across from its reverse) and determines the stability of the sequences to exist together. Furthermore, various Dna repair functions too as regulatory functions are based on base of operations pair complementarity. In biotechnology, the principle of base pair complementarity allows the generation of Dna hybrids between RNA and Deoxyribonucleic acid, and opens the door to modern tools such as cDNA libraries. While almost complementarity is seen between two split up strings of Deoxyribonucleic acid or RNA, it is likewise possible for a sequence to accept internal complementarity resulting in the sequence binding to itself in a folded configuration.
Deoxyribonucleic acid and RNA base of operations pair complementarity [edit]

Complementarity between 2 antiparallel strands of DNA. The top strand goes from the left to the right and the lower strand goes from the correct to the left lining them up.

Left: the nucleotide base pairs that tin can form in double-stranded Dna. Between A and T in that location are two hydrogen bonds, while in that location are three betwixt C and G. Right: two complementary strands of DNA.
Complementarity is accomplished past distinct interactions between nucleobases: adenine, thymine (uracil in RNA), guanine and cytosine. Adenine and guanine are purines, while thymine, cytosine and uracil are pyrimidines. Purines are larger than pyrimidines. Both types of molecules complement each other and can only base of operations pair with the opposing type of nucleobase. In nucleic acid, nucleobases are held together past hydrogen bonding, which but works efficiently between adenine and thymine and betwixt guanine and cytosine. The base complement A = T shares two hydrogen bonds, while the base pair Grand ≡ C has three hydrogen bonds. All other configurations betwixt nucleobases would hinder double helix formation. Dna strands are oriented in opposite directions, they are said to exist antiparallel.[1]
| Nucleic Acid | Nucleobases | Base complement |
| DNA | adenine(A), thymine(T), guanine(Chiliad), cytosine(C) | A = T, 1000 ≡ C |
| RNA | adenine(A), uracil(U), guanine(One thousand), cytosine(C) | A = U, Thou ≡ C |
A complementary strand of DNA or RNA may exist constructed based on nucleobase complementarity.[2] Each base of operations pair, A = T vs. Chiliad ≡ C, takes upwards roughly the same space, thereby enabling a twisted DNA double helix germination without any spatial distortions. Hydrogen bonding between the nucleobases besides stabilizes the Deoxyribonucleic acid double helix.[3]
Complementarity of DNA strands in a double helix make it possible to apply ane strand as a template to construct the other. This principle plays an important part in Deoxyribonucleic acid replication, setting the foundation of heredity past explaining how genetic information can be passed down to the next generation. Complementarity is also utilized in DNA transcription, which generates an RNA strand from a Deoxyribonucleic acid template.[4] In addition, homo immunodeficiency virus, a single-stranded RNA virus, encodes an RNA-dependent DNA polymerase (contrary transcriptase) that uses complementarity to catalyze genome replication. The reverse transcriptase can switch betwixt two parental RNA genomes past copy-choice recombination during replication.[5]
Dna repair mechanisms such as proof reading are complementarity based and allow for error correction during Dna replication past removing mismatched nucleobases.[1] In general, amercement in ane strand of Dna can exist repaired by removal of the damaged section and its replacement past using complementarity to re-create information from the other strand, equally occurs in the processes of mismatch repair, nucleotide excision repair and base excision repair.[6]
Nucleic acids strands may likewise form hybrids in which single stranded Deoxyribonucleic acid may readily anneal with complementary DNA or RNA. This principle is the ground of commonly performed laboratory techniques such as the polymerase chain reaction, PCR.[i]
2 strands of complementary sequence are referred to as sense and anti-sense. The sense strand is, generally, the transcribed sequence of DNA or the RNA that was generated in transcription, while the anti-sense strand is the strand that is complementary to the sense sequence.
Cocky-complementarity and hairpin loops [edit]

A sequence of RNA that has internal complementarity which results in it folding into a hairpin
Self-complementarity refers to the fact that a sequence of Deoxyribonucleic acid or RNA may fold back on itself, creating a double-strand like structure. Depending on how close together the parts of the sequence are that are cocky-complementary, the strand may class hairpin loops, junctions, bulges or internal loops.[1] RNA is more probable to grade these kinds of structures due to base pair binding not seen in DNA, such equally guanine bounden with uracil.[1]

A sequence of RNA showing hairpins (far right and far upper left), and internal loops (lower left structure)
Regulatory functions [edit]
Complementarity can be found between short nucleic acrid stretches and a coding region or an transcribed gene, and results in base pairing. These brusque nucleic acid sequences are usually found in nature and accept regulatory functions such as gene silencing.[ane]
Antisense transcripts [edit]
Antisense transcripts are stretches of non coding mRNA that are complementary to the coding sequence.[7] Genome wide studies accept shown that RNA antisense transcripts occur commonly within nature. They are generally believed to increase the coding potential of the genetic code and add together an overall layer of complexity to cistron regulation. And then far, it is known that 40% of the human being genome is transcribed in both directions, underlining the potential significance of opposite transcription.[8] It has been suggested that complementary regions between sense and antisense transcripts would allow generation of double stranded RNA hybrids, which may play an of import role in gene regulation. For example, hypoxia-induced gene 1α mRNA and β-secretase mRNA are transcribed bidirectionally, and it has been shown that the antisense transcript acts as a stabilizer to the sense script.[ix]
miRNAs and siRNAs [edit]

Formation and office of miRNAs in a cell
miRNAs, microRNA, are brusk RNA sequences that are complementary to regions of a transcribed cistron and have regulatory functions. Current research indicates that circulating miRNA may be utilized as novel biomarkers, hence show promising prove to be utilized in disease diagnostics ..[10] MiRNAs are formed from longer sequences of RNA that are cut free past a Dicer enzyme from an RNA sequence that is from a regulator factor. These short strands demark to a RISC complex. They match up with sequences in the upstream region of a transcribed factor due to their complementarity to human action as a silencer for the gene in iii ways. I is by preventing a ribosome from binding and initiating translation. 2 is past degrading the mRNA that the circuitous has bound to. And three is by providing a new double-stranded RNA (dsRNA) sequence that Dicer can human action upon to create more miRNA to find and degrade more copies of the gene. Pocket-sized interfering RNAs (siRNAs) are similar in function to miRNAs; they come from other sources of RNA, merely serve a like purpose to miRNAs.[ane] Given their brusque length, the rules for complementarity ways that they can still be very discriminating in their targets of choice. Given that there are 4 choices for each base in the strand and a 20bp - 22bp length for a mi/siRNA, that leads to more than 1×ten12 possible combinations. Given that the human being genome is ~three.1 billion bases in length,[11] this means that each miRNA should only observe a match once in the entire human genome by accident.
Kissing hairpins [edit]
Kissing hairpins are formed when a single strand of nucleic acid complements with itself creating loops of RNA in the form of a hairpin.[12] When two hairpins come up into contact with each other in vivo, the complementary bases of the two strands form up and brainstorm to unwind the hairpins until a double-stranded RNA (dsRNA) complex is formed or the complex unwinds back to ii separate strands due to mismatches in the hairpins. The secondary structure of the hairpin prior to kissing allows for a stable structure with a relatively fixed modify in free energy.[13] The purpose of these structures is a balancing of stability of the hairpin loop vs binding strength with a complementary strand. Too stiff an initial binding to a bad location and the strands volition not unwind chop-chop enough; too weak an initial binding and the strands will never fully form the desired complex. These hairpin structures permit for the exposure of enough bases to provide a strong plenty check on the initial binding and a weak enough internal binding to allow the unfolding once a favorable match has been found.[13]
---C 1000--- C One thousand ---C Thou--- U A C M G C U A C Chiliad Chiliad C A One thousand C Thousand A A A Thou C U A A U CUU ---CCUGCAACUUAGGCAGG--- A GAA ---GGACGUUGAAUCCGUCC--- Thou A U U U U U C U C G C G C C G C G A U A U G C G C ---G C--- ---Grand C--- Kissing hairpins meeting up at the superlative of the loops. The complementarity of the two heads encourages the hairpin to unfold and straighten out to become one flat sequence of ii strands rather than two hairpins.
Bioinformatics [edit]
Complementarity allows information constitute in DNA or RNA to exist stored in a single strand. The complementing strand can exist adamant from the template and vice versa as in cDNA libraries. This also allows for analysis, like comparing the sequences of two different species. Shorthands accept been adult for writing downward sequences when in that location are mismatches (ambiguity codes) or to speed up how to read the opposite sequence in the complement (ambigrams).
cDNA Library [edit]
A cDNA library is a collection of expressed DNA genes that are seen every bit a useful reference tool in gene identification and cloning processes. cDNA libraries are constructed from mRNA using RNA-dependent DNA polymerase contrary transcriptase (RT), which transcribes an mRNA template into Deoxyribonucleic acid. Therefore, a cDNA library can only contain inserts that are meant to be transcribed into mRNA. This process relies on the principle of DNA/RNA complementarity. The cease product of the libraries is double stranded Dna, which may be inserted into plasmids. Hence, cDNA libraries are a powerful tool in mod research.[1] [fourteen]
Ambiguity codes [edit]
When writing sequences for systematic biological science it may exist necessary to have IUPAC codes that hateful "whatever of the two" or "any of the iii". The IUPAC lawmaking R (any purine) is complementary to Y (any pyrimidine) and Yard (amino) to Yard (keto). W (weak) and Due south (strong) are usually non swapped[15] but accept been swapped in the past by some tools.[sixteen] W and S denote "weak" and "stiff", respectively, and bespeak a number of the hydrogen bonds that a nucleotide uses to pair with its complementing partner. A partner uses the same number of the bonds to make a complementing pair.[17]
An IUPAC lawmaking that specifically excludes one of the three nucleotides tin be complementary to an IUPAC code that excludes the complementary nucleotide. For instance, V (A, C or G - "not T") can be complementary to B (C, Chiliad or T - "non A").
| Symbol[18] | Description | Bases represented | ||||
|---|---|---|---|---|---|---|
| A | adenine | A | ane | |||
| C | cytosine | C | ||||
| G | one thousanduanine | 1000 | ||||
| T | thymine | T | ||||
| U | uracil | U | ||||
| Westward | weak | A | T | 2 | ||
| S | strong | C | K | |||
| Chiliad | amino | A | C | |||
| K | keto | G | T | |||
| R | purine | A | One thousand | |||
| Y | pyrimidine | C | T | |||
| B | not A (B comes after A) | C | G | T | 3 | |
| D | non C (D comes later on C) | A | G | T | ||
| H | non G (H comes after G) | A | C | T | ||
| V | not T (V comes after T and U) | A | C | G | ||
| N or - | any base (not a gap) | A | C | Chiliad | T | 4 |
Ambigrams [edit]
Specific characters may exist used to create a suitable (ambigraphic) nucleic acid note for complementary bases (i.e. guanine = b, cytosine = q, adenine = n, and thymine = u), which makes information technology is possible to complement unabridged Deoxyribonucleic acid sequences by just rotating the text "upside down".[19] For instance, with the previous alphabet, buqn (GTCA) would read as ubnq (TGAC, opposite complement) if turned upside downwards.
- qqubqnnquunbbqnbb
- bbnqbuubnnuqqbuqq
Ambigraphic notations readily visualize complementary nucleic acid stretches such every bit palindromic sequences.[20] This characteristic is enhanced when utilizing custom fonts or symbols rather than ordinary ASCII or even Unicode characters.[xx]
See as well [edit]
- Base of operations pair
References [edit]
- ^ a b c d due east f g h Watson, James, Cold Leap Harbor Laboratory, Tania A. Baker, Massachusetts Institute of Technology, Stephen P. Bong, Massachusetts Institute of Engineering, Alexander Gann, Cold Leap Harbor Laboratory, Michael Levine, University of California, Berkeley, Richard Losik, Harvard University ; with Stephen C. Harrison, Harvard Medical (2014). Molecular biological science of the gene (Seventh ed.). Boston: Benjamin-Cummings Publishing Company. ISBN978-0-32176243-6.
- ^ Pray, Leslie (2008). "Discovery of DNA structure and part: Watson and Crick". Nature Didactics. 1 (1): 100. Retrieved 27 November 2013.
- ^ Shankar, A; Jagota, A; Mittal, J (Oct 11, 2012). "DNA base dimers are stabilized by hydrogen-bonding interactions including non-Watson-Crick pairing well-nigh graphite surfaces". The Journal of Physical Chemistry B. 116 (xl): 12088–94. doi:10.1021/jp304260t. PMID 22967176.
- ^ Hood, L; Galas, D (Jan 23, 2003). "The digital code of Dna". Nature. 421 (6921): 444–8. Bibcode:2003Natur.421..444H. doi:10.1038/nature01410. PMID 12540920.
- ^ Rawson JMO, Nikolaitchik OA, Keele BF, Pathak VK, Hu WS. Recombination is required for efficient HIV-one replication and the maintenance of viral genome integrity. Nucleic Acids Res. 2018;46(20):10535-10545. DOI:10.1093/nar/gky910 PMID 30307534
- ^ Bit O, Nielsen O. DNA repair. J Cell Sci. 2004;117(Pt iv):515-517. DOI:10.1242/jcs.00952
- ^ He, Y; Vogelstein, B; Velculescu, VE; Papadopoulos, North; Kinzler, KW (Dec xix, 2008). "The antisense transcriptomes of homo cells". Science. 322 (5909): 1855–vii. Bibcode:2008Sci...322.1855H. doi:ten.1126/science.1163853. PMC2824178. PMID 19056939.
- ^ Katayama, S; Tomaru, Y; Kasukawa, T; Waki, K; Nakanishi, M; Nakamura, K; Nishida, H; Yap, CC; Suzuki, M; Kawai, J; Suzuki, H; Carninci, P; Hayashizaki, Y; Wells, C; Frith, Yard; Ravasi, T; Pang, KC; Hallinan, J; Mattick, J; Hume, DA; Lipovich, 50; Batalov, Southward; Engström, PG; Mizuno, Y; Faghihi, MA; Sandelin, A; Chalk, AM; Mottagui-Tabar, S; Liang, Z; Lenhard, B; Wahlestedt, C; RIKEN Genome Exploration Research Group; Genome Scientific discipline Group (Genome Network Projection Core Group); FANTOM Consortium (Sep 2, 2005). "Antisense transcription in the mammalian transcriptome". Scientific discipline. 309 (5740): 1564–six. Bibcode:2005Sci...309.1564R. doi:10.1126/science.1112009. PMID 16141073. S2CID 34559885.
- ^ Faghihi, MA; Zhang, M; Huang, J; Modarresi, F; Van der Brug, MP; Nalls, MA; Cookson, MR; St-Laurent G, tertiary; Wahlestedt, C (2010). "Show for natural antisense transcript-mediated inhibition of microRNA role". Genome Biological science. eleven (five): R56. doi:ten.1186/gb-2010-eleven-5-r56. PMC2898074. PMID 20507594.
- ^ Kosaka, N; Yoshioka, Y; Hagiwara, K; Tominaga, Northward; Katsuda, T; Ochiya, T (Sep v, 2013). "Trash or Treasure: extracellular microRNAs and cell-to-cell communication". Frontiers in Genetics. 4: 173. doi:x.3389/fgene.2013.00173. PMC3763217. PMID 24046777.
- ^ "Ensembl genome browser 73: Human being sapiens - Assembly and Genebuild". Ensembl.org . Retrieved 27 November 2013.
- ^ Marino, JP; Gregorian RS, Jr; Csankovszki, G; Crothers, DM (Jun 9, 1995). "Aptitude helix formation between RNA hairpins with complementary loops". Scientific discipline. 268 (5216): 1448–54. Bibcode:1995Sci...268.1448M. doi:10.1126/science.7539549. PMID 7539549.
- ^ a b Chang, KY; Tinoco I, Jr (May thirty, 1997). "The structure of an RNA "kissing" hairpin complex of the HIV TAR hairpin loop and its complement". Periodical of Molecular Biology. 269 (one): 52–66. doi:x.1006/jmbi.1997.1021. PMID 9193000.
- ^ Wan, KH; Yu, C; George, RA; Carlson, JW; Hoskins, RA; Svirskas, R; Stapleton, M; Celniker, SE (2006). "High-throughput plasmid cDNA library screening". Nature Protocols. i (2): 624–32. doi:10.1038/nprot.2006.ninety. PMID 17406289. S2CID 205463694.
- ^ Jeremiah Organized religion (2011), conversion tabular array
- ^ arep.med.harvard.edu A tool page with the note nigh the applied West-S conversion patch.
- ^ Opposite-complement tool page with documented IUPAC code conversion, source code bachelor.
- ^ Classification Committee of the International Spousal relationship of Biochemistry (NC-IUB) (1984). "Classification for Incompletely Specified Bases in Nucleic Acrid Sequences". Retrieved 2008-02-04 .
- ^ Rozak DA (2006). "The practical and pedagogical advantages of an ambigraphic nucleic acid note". Nucleosides Nucleotides Nucleic Acids. 25 (7): 807–13. doi:ten.1080/15257770600726109. PMID 16898419. S2CID 23600737.
- ^ a b Rozak, DA; Rozak, AJ (May 2008). "Simplicity, part, and legibility in an enhanced ambigraphic nucleic acid note". BioTechniques. 44 (6): 811–3. doi:10.2144/000112727. PMID 18476835.
External links [edit]
- Reverse complement tool
- Reverse Complement Tool @ Deoxyribonucleic acid.UTAH.EDU Archived 2018-08-29 at the Wayback Machine
richardsonknort1999.blogspot.com
Source: https://en.wikipedia.org/wiki/Complementarity_(molecular_biology)
0 Response to "what does it mean for dna to be complementary"
Post a Comment